Genetic markers
- Genes coding for the Immune system, Microbe infections, & Inflammatory system
- Mitochondria DNA and Gene tests
These are listed in the following paper
Mitochondrial DNA variants correlate with symptoms in myalgic encephalomyelitis/chronic fatigue syndrome.
J Transl Med. 2016 Jan 20;14(1):19. doi: 10.1186/s12967-016-0771-6. Billing-Ross P1, Germain A2, Ye K3, Keinan A4, Gu Z5, Hanson MR6.
- Early T‐cell activation‐1 gene and its gene product osteopontin
'An extensive monograph has described the early T‐cell activation‐1 gene and its gene product osteopontin paradigm.( Patarca‐Montero R. Chronic fatigue syndrome, genes, and infection: the eta‐1/Op paradigm. New York: Haworth Medical Press, 2003). This gene and its product have a vital role in initiating the response to infection by viruses (picornaviruses, herpes and HIV) and other microorganisms (including chlamydiae, coxiellae, rickettsiae and mycobacteria); autoimmune disease; cellular motility and communication; the regulation of phosphate and calcium metabolism, and bone growth and development; and numerous other body systems, including bone, joints and tendons, skin, kidney, heart, blood vessels, gastrointestinal system, lungs, central nervous, reproductive and auditory systems; and neoplasia. The widespread distribution and expression of the early T‐cell activation‐1 gene and its gene product osteopontin complex is consistent with the complex and extensive features of ME/CFS and provides a basis for a greater understanding of this illness.'
Myalgic encephalomyelitis: a review with emphasis on key findings in biomedical research.
M Hooper. J Clin Pathol. 2007 May; 60(5): 466–471.
doi: 10.1136/jcp.2006.042408
The Eta-1/Op Paradigm focuses on the Early T lymphocyte activation-1/osteopontin gene (Eta-1/Op), a cytokine that offers natural resistance to bacteria, and viruses that may play a role in the link between microbial infections and ME. This is proposed by geneticist Dr. Patarca-Montero who has stdied this fiels for many years. He provides scientific evidence in the following book - Chronic Fatigue Syndrome, Genes, and Infection: The ETA-1/OP Paradigm—What Does the Literature Say? . This genetic abnormality may be a factor in some ME subgroups predisposing them to immune system weaknesses and infections. . This genetic abnormality may be a factor in some ME subgroups predisposing them to immune system weaknesses and infections.
- The following research produced by Scottish researchers using DNA chip microarray technology
shows abormalities in 366 genes in ME/CFS patients. Of these, 286 genes were significantly up-regulated and 80 significantly down-regulated in patient samples. These genes code for immune system, microbe infections, inflammatory system, cell apoptosis and oxidative stress.
A gene signature for post-infectious chronic fatigue syndrome. John W Gow, Suzanne Hagan, Pawel Herzyk, Celia Cannon, Peter O Behan, and Abhijit Chaudhuri. BMC Med Genomics. 2009; 2: 38.
- Genes for Natural Killer Cells
- Genetic condition involving absence of NK cells in peripheral blood. This is a rare genetic condition involving three key genes (MCM4, FCGR3A, & GATA2)
- Genetic polymorphisms that result in deficient NK cells
- TLR4
- Light AR, White AT, Hughen RW, Light KC. Moderate exercise increases expression for sensory, adrenergic, and immune genes in chronic fatigue syndrome patients but not in normal subjects. J Pain. 2009;10:1099–112.
- White AT, Light AR, Hughen RW, Vanhaitsma TA, Light KC. Differences in metabolite-detecting, adrenergic, and immune gene expression after moderate exercise in patients with chronic fatigue syndrome, patients with multiple sclerosis, and healthy controls. Psychosom Med. 2012;74:46–54.
- Gow JW, Hagan S, Herzyk P, Cannon C, Behan PO, Chaudhuri A. A gene signature for post-infectious chronic fatigue syndrome. BMC Med Genomics. 2009;2:38.
- Other Neurological Ilnesses
- Nagyoszi P, Wilhelm I, Farkas AE, Fazakas C, Dung NT, Haskó J, Krizbai IA (2010) Expression and regulation of toll-like receptors in cerebral endothelial cells. Neurochem Int 57(5):556–64 168.
- Andersson A, Covacu R, Sunnemark D, Danilov AI, Dal Bianco A, Khademi M, Wallström E, Lobell A, Brundin L, Lassmann H, Harris RA (2008) Pivotal advance: HMGB1 expression in active lesions of human and experimental multiple sclerosis. J Leukoc Biol 84(5):1248–55 169.
- Bsibsi M, Ravid R, Gveric D, van Noort JM (2002) Broad expression of Toll-like receptors in the human central nervous system. J Neuropathol Exp Neurol 61(11):1013–21
- Kim C, Ho DH, Suk JE, You S, Michael S, Kang J, Joong Lee S, Masliah E, Hwang D, Lee HJ, Lee SJ (2013) Neuron-released oligomeric α-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat Commun 4:1562
- Tiffin N, Adeyemo A, Okpechi I (2013) A diverse array of genetic factors contribute to the pathogenesis of systemic lupus erythematosus. Orphanet J Rare Dis 8:2. doi:10.1186/1750-1172-8-2 173.
- Low HZ, Witte T (2011) Aspects of innate immunity in Sjögren’s syndrome. Arthritis Res Ther 13(3):218. doi:10.1186/ar3318 174.
- Goh FG, Midwood KS (2012) Intrinsic danger: activation of Tolllike receptors in rheumatoid arthritis. Rheumatology (Oxford) 51(1):7–23. doi:10.1093/rheumatology/ker257
- Wang PF, Fang H, Chen J, Lin S, Liu Y, Xiong XY, Wang YC, Xiong RP, Lv FL, Wang J, Yang QW (2014) Polyinosinicpolycytidylic acid has therapeutic effects against cerebral ischemia/reperfusion injury through the downregulation of TLR4 signaling via TLR3. J Immunol 192(10):4783–94. doi:10.4049/ jimmunol.1303108
- HLA abnormalities. Often associated with abnormal CD38 levels.
- 4 to 6 fold increased relative risk for DR4, DR3
and DQ3 (Keller et al, 1992)
- HLA DR haplotypes in 112 South Florida CFS
patients, compared to 5,000 regional and
national controls. (Klimas et al., 2007)
- Keller RH, Lane JL, Klimas N, Reiter WM, Fletcher MA, van Riel F et al. Association
between HLA class II antigens and the chronic fatigue immune dysfunction
syndrome. Clin Infect Dis 1994; 18: S154–S156.
- HLA-DR4 positive. Chronic Fatigue Syndrome. JA Goldstein. pub: The Chronic Fatigue Syndrome Institute, Beverly Hills, California 1990 ISBN 0-9625654-0-7
- HLA-DQA1. Association of chronic fatigue syndrome with human leucocyte antigen class II alleles. J Smith, E L Fritz, J R Kerr, A J Cleare, S Wessely, and D L Mattey. J Clin Pathol. 2005 August; 58(8): 860–863.
- HLA DR abnormalities. Chronic fatigue syndrome: clinical condition associated with immune activation. A.L. Landay, E.T. Lennette, C. Jessop, J.A. Levy. The Lancet, Volume 338, Issue 8769, 21 September 1991, Pages 707-712.
- Lloyd, A. R., D. Wakefield, C. R. Boughton, and J. M. Dwyer. 1989. Immunological abnormalities in the chronic fatigue syndrome. Med. J. Austral. 151:122-124.
- HLA-DQ1. The frequency of HLA Class II antigens in chronic fatigue syndrome. Schacterle, RS., Milford, EL and Komaroff, AL. Journal of Chronic Fatigue Syndrome, 2003, 11, 4, 33-42.
- Immunologic Abnormalities Associated with Chronic Fatigue Syndrome Edward Barker, Sue F. Fujimura, Mitchell B. Fadem, Alan L. Landay, and Jay A. Levy
- Genetic
Marker - HLA DR 1501. ' Of particular note is the high incidence of a genetic marker called
HLA DR 1501 in my fatigue sufferer population. This genetic marker is
four times more prevalent in my patient group than it is in a normal
background population. This marker is also present in almost every
patient with another fatigue condition known as narcolepsy, which is a
brain disorder that results in excessive daytime sleepiness, as well
as in other symptomatology. This finding of an increased prevalence of
HLA DR 1501 in my CFS population suggests that some of these patients
might also have narcolepsy, or a variant thereof. Alternatively, this
marker may point not only to a genetic predisposition to narcolepsy,
but perhaps to CFS as well. ' Dr John Whiting, Dr Philip Stowell and Dr Gary Deed, Newcastle
University, Australia.
- Published articles on Google Scholar
- T cells & Other Immune cells
- " 407 candidate autosomal SNPs were associated
with a diagnosis of ME/CFS in our cohorts (Po3.3 × 10 − 5
;
Supplementary Table 1) and 35 SNPs were identified on the X
chromosome. Twenty-three SNPs were significant at Po1.0 10 − 10
(Table 1, GWAS P-value). The most significant SNP (rs12235235,
genotypic association P = 5.76 × 10 − 16) was identified in the
intragenic region of the gene RECK (Reversion-Inducing
Cysteine-Rich Protein With Kazal Motifs), a putative negative
regulator of matrix metalloproteinases. In addition, among this
group of 23 SNPs, two were in the T-cell receptor alpha locus and
TRA (rs17255510 and rs11157573) and one in the T-cell receptor
alpha/delta locus (rs10144138). We also observed an SNP in the
intragenic region of the GRIK3 gene (rs3913434), a glutamate
neurotransmitter receptor and an ortholog of the GRIK2 gene,
which was identified as a highly statistically significant SNP in a
previous ME/CFS GWAS conducted in 2011 by Smith et al.
(genotypic association P-value P = 0.001 and P = 0.002. ) "
"The implications of multiple SNPs in the intragenic regions of TCA loci are more obvious. In the thymus, the TCA gene undergoes somatic recombination to give rise to diverse amino-acid sequences in the antigen-binding regions of the alpha chain of T-cell receptors. T-cell receptors recognize antigens bound to major histocompatibility complex class I and class II molecules and, therefore, are critical components of adaptive immunity. Major histocompatibility complex can present antigens from nearly all forms of pathogens; however, T cells that recognize 'self' antigens and have escaped negative selection in the thymus can promote autoimmune disease. Indeed, polymorphisms in the TCA locus have been described in association with autoimmune disease."
"therefore this
polymorphism produces a non-conservative substitution and
potentially may lead to decreased functionality of the receptor.
CLEC4M is a pattern recognition receptor capable of binding to a
broad range of pathogens, including hepatitis C virus,44 human
immunodeficiency virus45 and Mycobacterium tuberculosis.
49 A
dysregulation of CLEC4M may have significant consequences in
the pathogenesis of infectious diseases.50"
Genome-wide association analysis identifies genetic variations in subjects with myalgic encephalomyelitis/chronic fatigue syndrome KA Schlauch
, SF Khaiboullina, KL De Meirleir
, S Rawat
, J Petereit, AA Rizvanov, N Blatt, T Mijatovic
, D Kulick, A Palotás, and
VC Lombardi. Transl Psychiatry (2015) 6, doi:10.1038/tp.2015.208
- Sabath et al. reported that ME/CFS cases and
their respective twins displayed a trend of increased circulating
CD62L(+) T cells in several T-cell subsets
Sabath DE, Barcy S, Koelle DM, Zeh J, Ashton S, Buchwald D. Cellular immunity in
monozygotic twins discordant for chronic fatigue syndrome. J Infect Dis 2002; 185: 828–832
- Lymphocytes' Genes
- Identification of novel expressed sequences, up-regulated in the leucocytes of chronic fatigue syndrome patients.
Powell R, Ren J, Lewith G, Barclay W, Holgate S, Almond J.
Clin Exp Allergy. 2003 Oct;33(10):1450-6.
- Smith J, Fritz EL, Kerr JR, Cleare AJ, Wessely S, Mattey DL. Association of chronic
fatigue syndrome with human leucocyte antigen class II alleles. J Clin Pathol 2005;
58: 860–863.
- TNF-857 and IFNgamma 874 rare alleles
Carlo-Stella N, Badulli C, De Silvestri A, Bazzichi L, Martinetti M, Lorusso L et al. A
first study of cytokine genomic polymorphisms in CFS: Positive association of
TNF-857 and IFNgamma 874 rare alleles. Clin Exp Rheumatol 2006; 24: 179–182.
- DNA Methylation Modifications
- "We found an increased abundance of differentially methylated genes related to the immune response, cellular metabolism, and kinase activity. Genes associated with immune cell regulation, the largest coordinated enrichment of differentially methylated pathways, showed hypomethylation within promoters and other gene regulatory elements in CFS. These data are consistent with evidence of multisystem dysregulation in CFS and implicate the involvement of DNA modifications in CFS pathology."
DNA Methylation Modifications Associated with Chronic Fatigue Syndrome. Wilfred C. de Vega,
Suzanne D. Vernon,
Patrick O. McGowan. Plos One, August 11, 2014
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen S-E, et al. (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7: e41361. doi: 10.1371/journal.pone.0041361
- Genes, Infections and Immune Subgroups
- Microbial infections in eight genomic subtypes of chronic fatigue syndrome/myalgic encephalomyelitis. Lihan Zhang, John Gough, David Christmas, Derek L Mattey, Selwyn C M Richards, Janice Main, Derek Enlander, David Honeybourne, Jon G Ayres, David J Nutt, Jonathan R Kerr. J Clin Pathol 2010;63:156-164 doi:10.1136/jcp.2009.072561
- "In ME/CFS, there are three main abnormalities in gene expression studies; these involve
the
immune system, mitochondrial function and G
-
protein signalling. There are seven genes
upregulated in ME/CFS
–
those associated with apoptosis, pesticides, mitochondrial function,
demyelination and viral binding sites"
From presentations at the 8
th
International Association of Chronic Fatigue Syndrome (IACFS)
Conference, Fort Lauderdale, Florida, held on 10
–
14 January 2007:
J Kerr, St Georges, London
- According to Dr. Kerr and his research teams, there are 88 genes which are abnormally expressed and these correlate to physical dysfunctions and abnormalities and infections in ME patients. He also found 7 subgroups in ME. The 88 genes are detailed in the following document ; click on it to view it. Gene Expression Subtypes in Patients with Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. Jonathan R. Kerr, Robert Petty, Beverley Burke, John Gough, David Fear, Lindsey I. Sinclair, Derek L. Mattey, Selwyn C. M. Richards, Jane Montgomery, Don A. Baldwin, Paul Kellam, Tim J. Harrison, George E. Griffin, Janice Main, Derek Enlander, David J. Nutt and Stephen T. Holgate. The Journal of Infectious Diseases 2008; 197:1171– 84
- Differentially expressed genes in chronic fatigue
syndrome/myalgic encephalomyelitis (CFS/ME) patients reveal seven subtypes with
distinct clinical phenotypes. Petty R, Burke B, et al.. J Infect Dis 2008;197:1171e84
- Dr. Kerr's team identified 35 abnormal gene expressions through microarray and 16 through PCR. Gene expression in peripheral blood mononuclear cells (PBMC) of CFS patients. Kaushik N, Fear DJ, Richards SC, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DA, Holgate ST, Kerr JR. Journal of Clinical Pathology / Molecular Pathology 2005;58(8):826-32
- Kerr J R, Binns J H. Presentation to the Invest in ME Conference, London, 12 May 2006. DVDs of the presentations on can be obtained from http://www.investinme.org
- Genes and Chemical Poisoning
- NTE
The upregulated neuropathy target esterase, NTE, gene has an important role in neuronal function and is significant in cases of chemical poisoning or pesticide poisoning.
Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome.
Kaushik N, Fear D, Richards SC, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DA, Holgate ST, Kerr JR
J Clin Pathol. 2005 Aug; 58(8):826-32.
Kerr J R, Binns J H. Presentation to the Invest in ME Conference, London, 12 May 2006. DVDs of the presentations on can be obtained from http://www.investinme.org
Scientific progress in understanding Gulf War veterans' illnesses: report and recommendations of the Research Advisory Committee on Gulf War Veterans' Illnesses, RACGWI, November 2004. http://www1.va.gov/rac-gwvi/docs/ReportandRecommendations_2004.pdf
- Post Exercise Changes to Genes in ME/CFS
Abnormally changed genes associated with the following biological pathways and proteins after exercise have been linked to CFS according to studies conducted by Dr. Light in the University of Utah, USA:
1. sensory receptors (ASICS, 2PX4, 2PX5, TPRIV1)
2. adrenergic receptors (sympathetic nervous system) (Alpha 2a, Beta-1, Beta-2, COMT)
3. cytokine receptors (IL-6, IL-10, TNF-a, CD14, TLRF4)
There is significant and continuing abnormalities in these genes and receptors after exercise.according to studies conducted by Dr. Light in Utah. See Scientific Evidence Section - Genetic Markers
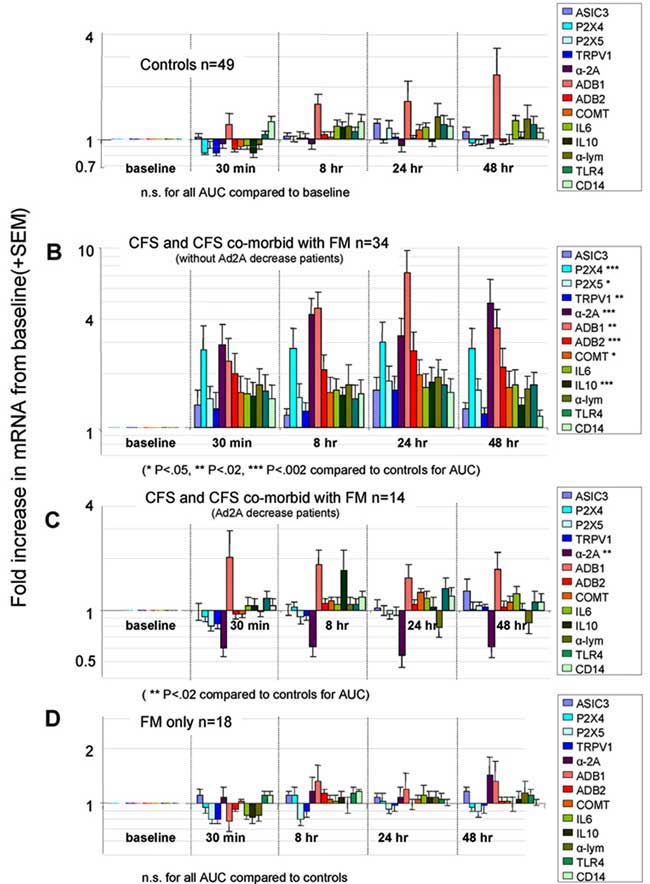
Source: Moderate Exercise Increases Expression for Sensory, Adrenergic, and Immune Genes in Chronic Fatigue Syndrome Patients But Not in Normal Subjects
Alan R. Lightemail address
,
Andrea T. White
,
Ronald W. Hughen
,
Kathleen C. Light. The Journal of Pain
Volume 10, Issue 10 , Pages 1099-1112, October 2009.
- Genome-wide association studies (GWAS)
- Two SNPs in GRIK2, a gene implicated in a number of
neurological maladies such as autism and schizophrenia and an
SNP within the NPAS2 gene, which is a putative circadian
clock gene.
Smith AK, Fang H, Whistler T, Unger ER, Rajeevan MS. Convergent genomic studies
identify association of GRIK2 and NPAS2 with chronic fatigue syndrome. Neuropsychobiology 2011; 64: 183–194
- " 407 candidate autosomal SNPs were associated
with a diagnosis of ME/CFS in our cohorts (Po3.3 × 10 - 5
;
Supplementary Table 1) and 35 SNPs were identified on the X
chromosome. Twenty-three SNPs were significant at Po1.0 10 - 10
(Table 1, GWAS P-value). The most significant SNP (rs12235235,
genotypic association P = 5.76 × 10 - 16) was identified in the
intragenic region of the gene RECK (Reversion-Inducing
Cysteine-Rich Protein With Kazal Motifs), a putative negative
regulator of matrix metalloproteinases. In addition, among this
group of 23 SNPs, two were in the T-cell receptor alpha locus and
TRA (rs17255510 and rs11157573) and one in the T-cell receptor
alpha/delta locus (rs10144138). We also observed an SNP in the
intragenic region of the GRIK3 gene (rs3913434), a glutamate
neurotransmitter receptor and an ortholog of the GRIK2 gene,
which was identified as a highly statistically significant SNP in a
previous ME/CFS GWAS conducted in 2011 by Smith et al.
(genotypic association P-value P = 0.001 and P = 0.002. ) "
"Both GRIK2 and GRIK3 code
for transmembrane subunits of neuroexcitatory receptors, belonging
to the kainate family of glutamate receptors. These receptors
are composed of four subunits and function as ligand-activated
ion channels on presynaptic and postsynaptic neurons"
Genome-wide association analysis identifies genetic variations in subjects with myalgic encephalomyelitis/chronic fatigue syndrome KA Schlauch
, SF Khaiboullina, KL De Meirleir
, S Rawat
, J Petereit, AA Rizvanov, N Blatt, T Mijatovic
, D Kulick, A Palotás, and
VC Lombardi. Transl Psychiatry (2015) 6, doi:10.1038/tp.2015.208
- Use of single-nucleotide polymorphisms (SNPs) to distinguish gene expression subtypes of chronic fatigue syndrome/myalgic encephalomyelitis (CFS/ME). Nana Shimosako, Jonathan R Kerr. J Clin Pathol 2014;67:1078-1083 doi:10.1136/jclinpath-2014-202597
- Abnormal Genes and Genetic Pathways
The following video of a lecture by Dr. Gordon Broderick, University of Alberta presents the immune system findings and underlying genes involved in this illness
Dynamic Modeling for ME/CFS & Gulf War Illness Interventions from ME-CFSCommunity.com on Vimeo.
Dr. Gordon Broderick's research team have uncovered several abnormal gene pathways which have direct effects on immune system function, HPA axis function, neurological function and metabolic function. These can differentiate healthy people from those with ME/CFS and also can differentiate ME/CFS from GWS patients.
1. Suppression of alanine and aspartate metabolism (kegg)
2.
Starch and Glucose metabolism (kegg) - increased
3.
Glycolysis / Gluconeogenesis (kegg) - increased
4. Phenylalanine metabolism (kegg) - decreased
5.
Pentose Phosphate pathway (kegg) - increased
6.
Trk (Tyrosine Kinase) receptor signalling mediated by pi3K and plc-gamma (nci/nature) - decreased
7. Disengagement of growth factor signalling and tissue repair ; aurora A
signalling decreased.
Source: Presentation of scientific research findings of Dr. Gordon Broderick
Prefential Pathway Activation in Gulf War Veterans with Unexplained Neuroendocrine Immune Imbalances. G. Broderick et al. (2011)
- Genes and Subgroups
- According to Dr. Kerr and his research teams, there are 88 genes which are abnormally expressed and these correlate to physical dysfunctions and abnormalities and infections in ME patients. He also found 7 subgroups in ME. The 88 genes are detailed in the following document ; click on it to view it. Gene Expression Subtypes in Patients with Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. Jonathan R. Kerr, Robert Petty, Beverley Burke, John Gough, David Fear, Lindsey I. Sinclair, Derek L. Mattey, Selwyn C. M. Richards, Jane Montgomery, Don A. Baldwin, Paul Kellam, Tim J. Harrison, George E. Griffin, Janice Main, Derek Enlander, David J. Nutt and Stephen T. Holgate. The Journal of Infectious Diseases 2008; 197:1171– 84
- Dr. Kerr's team identified 35 abnormal gene expressions through microarray and 16 through PCR. Gene expression in peripheral blood mononuclear cells (PBMC) of CFS patients. Kaushik N, Fear DJ, Richards SC, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DA, Holgate ST, Kerr JR. Journal of Clinical Pathology / Molecular Pathology 2005;58(8):826-32
- Microbial infections in eight genomic subtypes of chronic fatigue syndrome/myalgic encephalomyelitis. Lihan Zhang, John Gough, David Christmas, Derek L Mattey, Selwyn C M Richards, Janice Main, Derek Enlander, David Honeybourne, Jon G Ayres, David J Nutt, Jonathan R Kerr. J Clin Pathol 2010;63:156-164 doi:10.1136/jcp.2009.072561
- Use of single-nucleotide polymorphisms (SNPs) to distinguish gene expression subtypes of chronic fatigue syndrome/myalgic encephalomyelitis (CFS/ME). Nana Shimosako, Jonathan R Kerr. J Clin Pathol 2014;67:1078-1083 doi:10.1136/jclinpath-2014-202597
- Chromosonal damage in most ME/CFS patients
Dr. Heng of Wayne State University (USA) found serious chromosone breakage in 100% of ME/CFS patients using spectral karyotyping. 53% were identified as having chromosome translocations, often considered to be a recognized stepping stone of the cancer initiation process. Details provided on the NCF web site at http://www.ncf-net.org/PressReleases.htm#nfc
- Test for chromosone damage using spectral karyotyping equipment used by Dr. Heng
- Test for internal radionuclides in patients. These would be sources of regular exposure to radiation, capable of causing chromosone damage. Some good research articles concerning radiactive particles in the
environment and food chain and the development of Cancers, immune system, endocrine and neurological illnesses can be found at http://www.ncf-net.org/radiation.htm
- Neuronal tryptophan hydroxylase (TPH2), catechol-O-methyltransferase (COMT) and nuclear receptor subfamily 3, group C, member 1 glucocorticoid receptor (NR3C1)
Combinations of single nucleotide polymorphisms in neuroendocrine effector and receptor genes predict chronic fatigue syndrome.
Goertzel BN1, Pennachin C, de Souza Coelho L, Gurbaxani B, Maloney EM, Jones JF.
- 5-HT transporter gene polyphormism in CFS patients.
Indication of endocrine abnormality induced by dysfunction of 5-HT system in CFS. Dysfunction in 5-HT system in the animal models. Reduced binding potential of 5-HTT in CFS patients
Neuroscience of Fatigue and CFS/ME by Using PET Molecular Imaging and Functional Neuroimaging
- Yasuyoshi Watanabe, Masaski Tanaka, Kei Mizuno, Akira Ishii, Emi Yamano, Sanae Fukuda, Yasuhito Nakatomi, Kouzi Yamaguti, and Hirohiko Kuratsune. IACFS/ME Conference. Translating Science into Clinical Care. March 20-23, 2014 • San Francisco, California, USA
- Association between serotonin transporter gene polymorphism and chronic fatigue syndrome
Masaaki Narita,
Naoko Nishigami,
Naoko Narita,
Kouzi Yamaguti,
Nobuo Okado,
Yasuyoshi Watanabe, Hirohiko Kuratsune. Biochemical and Biophysical Research Communications
Volume 311, Issue 2, 14 November 2003, Pages 264–266
- Genetic Inheritability
- Seattle CFS Cooperative Research Center Twin
study - genetic predisposition, hereditability
estimate of 51% (2nd World Conf); similar
results in Sweden, Australian studies
- MTHFR gene mutation found in some ME and CFS patients
The Human Genome Project found that this gene has been found to be mutated or abnormal in many people. Some ME and CFS patients have this genetic abnormality. This causes methylation cycle blockage, immune system abnormalities, thyroid and endocrine abnormalities, excess inflammation, antioxidant deficiencies, high homocysteine levels, arteriosclerosis and cardiac abnormalities, fatty liver, increased Cancer risk, neurotransmitter abnormalities and mood disorders, IBS, detoxification deficiency, multiple chemical sensitivity, cell growth and maintenance abnormalities. All of these are commonly found in ME and CFS patients.
Testing Labs
- DNA Methylation Pathway with Methylation Pathway Analysis
Some more info
http://www.stopthethyroidmadness.com/mthfr/
MTHFR Awareness
Google listings
Source: Video Lecture by doctor
- Abnormalities in Genes Regulating the HPA Axis
- A. K. Smith, P.D. White, E. Aslakson, U. Vollmer-Conna and M.S. Rajeevan, "Polymorphisms in Genes Regulating the HPA Axis Associated with Empirically Delineated Classes of Unexplained Chronic Fatigue," Pharmacogenomics, Vol. 7, 2006, pp. 387-394.
- J. R. Kerr, R. Petty, B. Burke, J. Gough, D. Fear, L. I. Sinclair, D. L. Mattey, S. C. Richards, J. Montgomery, D. A. Baldwin, P. Kellam, T. J. Harrison, G. E. Griffin, J. Main, D. Enlander, D. J. Nutt and S. T. Holgate, "Gene Expression Subtypes in Patients with Chronic Fatigue Syndrome/Myalgic Encephalomyelitis," Journal of Infectious Diseases, Vol. 197, No. 8, 2008, pp. 1171-1184.
- T. Saiki, T. Kawai, K. Morita, M. Ohta, T. Saito, K. Rokutan and N. Ban, "Identification of Marker Genes for Differential Diagnosis of Chronic Fatigue Syndrome," Molecular Medicine, 2008. PMID: 18596870
- M. S. Rajeevan, A. K. Smith, I. Dimulescu, E. R. Unger, S. D. Vernon, C. Heim, W. C. Reeves, "Glucocorticoid Receptor Polymorphisms and Haplotypes Associated with Chronic Fatigue Syndrome," Genes, Brain,& Behavior, Vol. 6, 2006, 167-176
- Jason LA, Sorenson M, Porter N, Belkairous N (2010), "An Etiological Model for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome", Neuroscience & Medicine, 2011, 2, 14-27, PMID: 21892413
- L. A. Jason, M. Sorenson, N. Porter, M. Brown, A. Lerch, C. Van der Eb and J. Mikovits, "Possible Genetic Dysregulation in Pediatric CFS," Psychology, Vol. 1, 2010, pp. 247-251.
- CNDP1 gene abnormality
Baraniuk et al. believe that patients with ME/CFS have a longer rather than shorter version of the gene CNDP1
(carnosine dipeptidase).A chronic fatigue syndrome - related proteome in human cerebrospinal fluid. Baraniuk, JN., Casado, B., Maibach, H., Clauw, DJ., Pannell, LK and Hess, S. BMC Neurology, 2005, 5:22.
- DQB1*0602 marker
A. R. Spitzer and M. Broadman, "Treatment of the Narcoleptiform Sleep Disorder in Chronic Fatigue Syndrome and Fibromyalgia with Sodium Oxybate," Pain Practice, Vol. 10, No. 1, 2010, pp. 54-59.
- Further abnormal changes in genes have been identified for the following in ME/CFS:
- Monoamine oxidase A (MAO A)
- Monoamine oxidase B (MAO B)
- Serotonin transporter (5-HTT) gene promoter
- Corticosteroid binding globulin (CBG)
- Tumor necrosis factor (TNF)
- Interferon gamma (IFN-gamma)
- Proopiomelanocortin (POMC)
- Ehlers-Danlos Syndrome (EDS)
EDS is
found in some ME and CFS patients.
An EDS specialist will need to carry out diagnosis based on
(i) symptoms
(ii) Brighton criteria and Brighton Score
(iii) genetic tests
(iv) collagen typing via skin biopsy
(v) echocardiogram, and lysyl hydroxylase or oxidase activity.
The abnormal genes known to be involved are : COL3A1, TNXB, COL5A1, COL5A2, COL1A1, PLOD1, COL1A1, COL1A2, ADAMTS2, unspecified gene, locus 12p13, unspecified gene, locus 2q34, Beasley–Cohen type (rare), B4GALT7, Tenascin-X deficiency – TNXB, D4ST1-Deficient Ehlers–Danlos syndrome (Adducted Thumb-Clubfoot Syndrome) CHST14.
Further Research Information
Ehlers-Danlos Syndrome, Hypermobility Type: An Underdiagnosed Hereditary Connective Tissue Disorder with Mucocutaneous, Articular, and Systemic Manifestations
- Moderate Exercise Increases Expression for Sensory, Adrenergic, and Immune Genes in Chronic Fatigue Syndrome Patients But Not in Normal Subjects. Alan R. Light et al. The Journal of Pain. Volume 10, Issue 10, October 2009, Pages 1099–1112
- Gene expression in peripheral blood mononuclear cells (PBMC) of CFS patients. Kaushik N, Fear DJ, Richards SC, McDermott CR, Nuwaysir EF, Kellam P, Harrison TJ, Wilkinson RJ, Tyrrell DA, Holgate ST, Kerr JR. Journal of Clinical Pathology / Molecular Pathology 2005;58(8):826-32
-
Differentially expressed genes in Chronic Fatigue Syndrome / Myalgic Encephalomyelitis (CFS/ME) patients reveal seven subtypes with distinct clinical phenotypes. Kerr JR, Petty R, Burke B, Gough J, Fear D, Sinclair LI, Mattey DL, Richards SC, Montgomery J, Baldwin DA, Kellam P, Harrison TJ, Griffin GE, Main J, Enlander D, Nutt DJ, Holgate ST. Journal of Infectious Diseases 2008;197:1171-84.
- Seven genomic subtypes of Chronic Fatigue Syndrome / Myalgic Encephalomyelitis (CFS/ME): a detailed analysis of gene networks and clinical phenotypes. Kerr JR, Burke B, Petty R, Gough J, Fear D, Mattey DL, Axford JS, Dalgleish AG, Nutt DJ. Journal of Clinical Pathology 2008;61:730-9
- Gene Expression Subtypes in Patients with Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. Jonathan R. Kerr, Robert Petty, Beverley Burke,
John Gough, David Fear, Lindsey I. Sinclair, Derek L. Mattey, Selwyn C. M. Richards, Jane Montgomery, Don A. Baldwin, Paul Kellam, Tim J. Harrison, George E. Griffin, Janice Main, Derek Enlander, David J. Nutt and Stephen T. Holgate. The Journal of Infectious Diseases 2008; 197:1171– 84
-
Gene profiling of patients with chronic fatigue syndrome/myalgic encephalomyelitis. Kerr JR. Curr Rheumatol Rep. 2008 Dec;10(6):482-91.
- J.W. Gow, C. Cannon, W.M.H. Behan, P. Herzyk, S. Keir,
G. Riboldi-Tunnicliffe, et al., "Whole-Genome (33,000
genes) Affymetrix DNA Microarray Analysis of Gene
Expression in Chronic Fatigue Syndrome," Paper presented
at the International Conference on Fatigue Science,
Karuizawa, Japan, 2005.
- Gow JW, Hagan S, Herzyk P, Cannon C, Behan PO, Chaudhuri A. A gene signature for
post
-
infectious chronic fatigue syndrome.
BMC Medl Genomics
(
2009
)
2
:
38. doi:
10.1186/1755
-
8794
-
2
-
38.
- Kaushik N, Fear D, Richards SC, McDer
mott CR, Nuwaysir EF, Kellam P, et al. Gene
expression in peripheral blood mononuclear cells from patients with chronic fatigue
syndrome.
J Clin Pathol
(
2005
)
58
:
826
-
832. doi: 10.1136/jcp.2005.025718.
- Fuite J, Vernon SD, Broderick G. Neuroendocrine and immune network re
-
modeling in
chronic fatigue syndrome: an exploratory analysis.
Genomics
(
2008
)
92
:
393
-
399. doi:
10.1016/j.ygeno.2008.08.008.
- Carlo-Stella N, Badulli C, De Silvestri A, Bazzichi L, Martinetti M, Lorusso L, Bombardieri S, Salvaneschi L, Cuccia M: A first study of cytokine genomic polymorphisms in CFS: positive association of TNF-857 and IFN-gamma 874 rare alleles. Clin Exp Rheumatol 2006, 24:179-182.
- T. Saiki, T. Kawai, K. Morita, M. Ohta, T. Saito, K. Rokutan and N. Ban, "Identification of Marker Genes for Differential Diagnosis of Chronic Fatigue Syndrome," Molecular Medicine, 2008. PMID: 18596870
- M. S. Rajeevan, A. K. Smith, I. Dimulescu, E. R. Unger, S. D. Vernon, C. Heim, W. C. Reeves, "Glucocorticoid Receptor Polymorphisms and Haplotypes Associated with Chronic Fatigue Syndrome," Genes, Brain,& Behavior, Vol. 6, 2006, 167-176.
- Aspler AL, Bolshin C, Vernon SD, Broderick G (2008) Evidence of inflammatory immune signaling in chronic fatigue syndrome: A pilot study of gene expression in peripheral blood. Behav Brain Funct 4: 44
-
Helbig K, Harris R, Ayres J, Dunckley H, Lloyd A, Robson J, Marmion BP: Immune response genes in the post-Q-fever fatigue syndrome, Q fever endocarditis and uncomplicated acute primary Q fever. QJM 2005, 98:565-574.
- A Twin Study of Chronic Fatigue.
Buchwald, Dedra MD; Herrell, Richard PhD; Ashton, Suzanne BS; Belcourt, Megan BS; Schmaling, Karen PhD; Sullivan, Patrick MD; Neale, Michael PhD, and; Goldberg, Jack PhD. Psychosomatic Medicine:
November/December 2001 - Volume 63 - Issue 6 - pp 936-943
-
Vollmer-Conna U, Piraino BF, Cameron B, Davenport T, Hickie I, Wakefield D, Lloyd AR, Dubbo Infection Outcomes Study Group: Cytokine polymorphisms have a synergistic effect on severity of the acute sickness response to infection. Clin Infect Dis 2008, 47:1418-1425
- Identification of marker genes for differential diagnosis of chronic fatigue syndrome.
Saiki T, Kawai T, Morita K, Ohta M, Saito T, Rokutan K, Ban N. Mol Med. 2008 Sep-Oct;14(9-10):599-607.
- Current research priorities in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis (CFS/ME):disease mechanisms, a diagnostic test and specific treatments. Jonathan R Kerr, Peter Christian, Andrea Hodgetts, Paul R Langford, Lakshmi D Devanur, Robert Petty, Beverley Burke, Lindsey I Sinclair, Selwyn C M Richards, Jane Montgomery, Clare McDermott, Tim J Harrison, Paul Kellam, David J Nutt and Stephen T Holgate. Journal of Clinical Pathology 2007;60(2):113-6.
- Single nucleotide polymorphisms (SNPs) associated with symptomatic infection and differential human gene expression in normal seropositive persons each implicate the cytoskeleton, integrin signalling and oncosuppression in the pathogenesis of human parvovirus B19 infection. Kerr JR, Kaushik N, Fear DJ, Baldwin D, Nuwaysir EF, Adcock IM. Journal of Infectious Diseases 2005;192(2):276-86.
- V. R. Falkenberg, B.M. Gurbaxani, E. R. Unger and M. S. Rajeevan, "Functional Genomics of Serotonin Receptor 2a (HTR2A): Interaction of Polymorphism, Methylation, Expression and Disease Association," Neuromolecural Medicine, 2010.
- Cytokine gene polymorphisms associated with symptomatic parvovirus B19 infection. Kerr JR, McCoy M, Burke B, Mattey DL, Pravica V, Hutchinson IV. J Clin Pathol. 2003 Oct;56(10):725-7.
- Microbial infections in eight genomic subtypes of Chronic Fatigue Syndrome / Myalgic Encephalomyelitis (CFS/ME)
Lihan Zhang, John Gough, David Christmas, Derek L Mattey, Selwyn CM Richards, Janice Main, Derek Enlander, David Honeybourne, Jon G Ayres, David J Nutt, Jonathan R Kerr. 2009
-
Broderick, G., Craddock, R., Whistler, T., Taylor, R., Klimas, N. and E. Unger. 2006. Identifying illness parameters using classical projection methods. Pharmacogenomics 7, 407-416.
- Integration of gene expression, clinical, and epidemiologic data to characterize Chronic Fatigue Syndrome.
Toni Whistler, Elizabeth R Unger, Rosane Nisenbaum and Suzanne D Vernon. Journal of Translational Medicine 2003, 1:10
- Fang, H., Xie, Q., Boneva, R., Fostel, J., Perkins, R. and W. Tong. 2006. Gene Expression profile exploration of a large dataset on chronic fatigue syndrome. Pharmacogenomics 7. 429-440.
- Evidence for a heritable predisposition to Chronic Fatigue Syndrome. Frederick Albright1, Kathleen Light, Alan Light, Lucinda Bateman and Lisa A Cannon-Albright. BMC Neurology 2011, 11:62
- Whistler, T., Taylor, R., Craddock, R., Broderick, G., Klimas, N and E. Unger. 2006. Gene expression correlates of unexplained fatigue. Pharmacogenomics 7. 395-405.
- Whistler T, Jones JF, Unger
ER, Vernon SD.
Exercise responsive genes measured in
peripheral blood of women with chronic fatigue syndrome and matched control
subjects.
BMC Physiol
(
2005
)
5
: 5.
- Goertzel, B., Pennachin, C., Coelho, L., Gurbaxani, B., Maloney, E. and J. Jones. 2006. Combinations of single nucleotide polymorphisms in neuroendocrine effector and receptor genes predict chronic fatigue syndrome. Pharmacogenomics 7, 475-83.
- A BIOMARKER FOR CFS FOUND? Presson, A., Sobel, E., Papp, J., Lusis, A., and S. Horvath. 2006. Integration of genetic and genomic approaches for the analysis of Chronic Fatigue Syndrome. THE 2006 CAMDA CONFERENCE.
- A Key Biological Pathway Altered in CFS ? Emmert-Streib, F., Glynn, E., Seidel, C., Bausch, C. and A. Mushegian. Detecting pathological pathways of the Chronic Fatigue Syndrome by the comparison of networks. THE 2006 CAMDA CONFERENCE
-
Evidence for a heritable predisposition to Chronic Fatigue Syndrome. Frederick Albright, Kathleen Light, Alan Light, Lucinda Bateman and Lisa A Cannon-Albright. BMC Neurology 2011, 11:62
- A. K. Smith, P.D. White, E. Aslakson, U. Vollmer-Conna and M.S. Rajeevan, "Polymorphisms in Genes Regulating the HPA Axis Associated with Empirically Delineated Classes of Unexplained Chronic Fatigue," Pharmacogenomics, Vol. 7, 2006, pp. 387-394.
- Jason LA, Sorenson M, Porter N, Belkairous N (2010), "An Etiological Model for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome", Neuroscience & Medicine, 2011, 2, 14-27, PMID: 21892413
- L. A. Jason, M. Sorenson, N. Porter, M. Brown, A. Lerch, C. Van der Eb and J. Mikovits, "Possible Genetic Dysregulation in Pediatric CFS," Psychology, Vol. 1, 2010, pp. 247-251.
- Smith AK, Fang H, Whistler T, Unger ER, Rajeevan MS (2011) Convergent genomic studies identify association of GRIK2 and NPAS2 with chronic fatigue syndrome. Neuropsychobiology 64: 183–194.
-
Dr.
Paul Cheney, North Carolina, research paper presented to the AACFS 5th
International Research, Clinical and Patient Conference, 2001 "
All chronic diseases studied including ME / CFS show prominent
circulating plasma RNA's not observed in normal controls. Prominent
RNA bands so far sequenced show homology with human genes which are
noted for their tendency for gene rearrangment under severe
physiologic stress. The sequences appear to be disease specific,
arying by less than 1% between individuals with similar illness. Gene
probes may therefore provide novel diagnostic and treatment
possibilities.
-
Polygenic
Contributions of Procoagulant Genetic Factors in Chronic Fatigue
Syndrome and Related Chronic Illnesses
Genetics in Medicine (2002)
|

